- HOME
- Products & Services
- Application Selection Guide
- Repeat Motif Detection
-
Oligonucleotide API CDMO
-
Custom DNA/RNA Systhesis
-
Next Generation Sequencing
- Next Generation Sequencing
-
Application Selection Guide
- Application Selection Guide
- Human Genome Sequencing
- Whole Genome Sequencing (Non-Human)
- Microbial genome sequencing
- Small scale sequencing (NGS Petit)
- GRAS-Di® Genotyping
- Repeat Motif Detection
- Library Prep for Challenging DNA Samples
- Gene Expression Analysis (Reference-Based)
- Isoform Sequencing (full-length mRNA-seq)
- De novo Transcriptome Sequencing
- Small RNA-Seq
- Microbial Community Analysis
- Shotgun Metagenomic Sequencing
- Metatranscriptome Sequencing
- ChIP-Seq
- CRISPR Screening
- Illumina Amplicon Sequencing
- PacBio Amplicon Sequencing
- Standard Pipeline Data Analysis
- Custom Data Analysis
- Gene Analysis from Pathological Specimens
- Sequencer Models and Sequencing Principles
- Sample Requirements
- How to Order
- Scientific Publications
-
Custom DNA Sequencing
-
Custom DNA Microarray
-
Protein Related Service
-
Laboratory Tools and Service
- Nucleic Acid Extraction
- RNA Purification
- RNA Quality Control
- Custom Measurement for synthetic DNA Molecular Weight
- Full-length cDNA library Production Service
- Stock Series
- LaboPass™ Products Series
- Koro Koro Rack
- Ukkie
- 8-tube strip
- Rhodococcus erythropolis Protein Expression System
- LipoTrust™ Series
Repeat Motif Detection
Overview
Repeat Motif Detection service identifies repeat motifs (e.g., microsatellites, SSRs, STRs) in genomic data using next-generation sequencing. Even for large plant and animal genomes, repeat motifs can be detected from a small amount of sequencing data.
About Repeat Motifs:
Repeat motifs are short, repeated sequences found in genomes and are often used as markers for polymorphism analysis. These are also referred to as microsatellites, simple sequence repeats (SSRs), or short tandem repeats (STRs).
Workflow
・ DNA library preparation
・ MiSeq 300 bp paired-end sequencing (100,000 read pairs or 60 Mb/sample)
・ Detection of repeat motifs
Price
201,000 JPY / sample (excluding tax)
Delivered Data
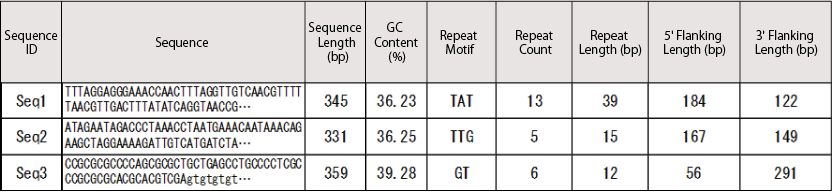
You will receive a comprehensive table summarizing repeat motifs and the flanking sequences, allowing you to design PCR primers for these regions.
Sample Requirements
| Sample Type | Total Amount | Concentration | Volume |
| DNA | ≥ 4 µg | ≥ 40 ng/µL | ≥ 30 µL |