ChIP-Seq
Overview
Chromatin Immunoprecipitation Sequrncing (ChIP-Seq) enables comprehensive detection of transcription factor binding sites and histone modification regions across the genome.
Workflow
推奨データ取得量
| Sequencing Specification | Illumina 150PE |
| Data Amount | 20 million read pairs per sample |
Data Analysis
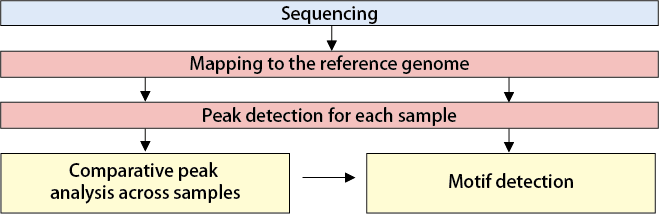
【Mapping and Peak Detection】
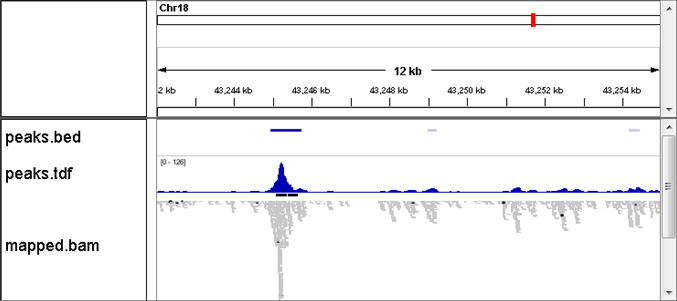
Reads are mapped to the reference genome, and peak regions are identified.
[Mapping Results]
【Comparative Analysis of Peak Regions Across Samples】
A comparison table is generated for peak regions across multiple samples, allowing identification of common peaks or peaks specific to a single sample.
【Motif Detection】
Highly frequent sequence motifs are identified from nucleotide sequences of genome-wide peak regions.
[Sequence logo representation]

Sample Requirements
| Sample Type | Total Amount | Concentration | Volume |
| ChIP-DNA | ≥ 20 ng | ≥ 1 ng/µL | ≥ 30 µL |
Frequently Asked Questions
We primarily offer shared analyses using Illumina 150bp paired-end sequencing.
For ChIP-Seq analysis, control samples such as input DNA or IgG-ChIP DNA are strongly recommended to remove background noise.
Input DNA Samples
Input DNA refers to fragmented genomic DNA samples without immunoprecipitation (IP) treatment. Peaks arising from biases during DNA fragmentation or PCR amplification can be used as background data.
IgG ChIP Samples
IgG ChIP samples provide information on non-specific binding to IgG. If the DNA recovery is insufficient for sequencing due to minimal non-specific binding, input DNA should be provided as a control instead.
