- HOME
- Products & Services
- Application Selection Guide
- Shotgun Metagenomic Sequencing
Shotgun Metagenomic Sequencing
Overview
Shotgun metagenomic sequencing involves creating a comprehensive sequencing library from genomic DNA extracted from environmental samples containing various microorganisms.
This method provides detailed insights into the microbial species present in the sample and the functional genes they harbor. Additionally, it allows for sequencing DNA from unculturable microorganisms and viruses.
Workflow
Specifications
| Platform | Illumina NovaSeq 6000 |
| Read Method | Paired-End (150 bp x 2) |
| Data Amount | 6 Gb/sample (20 million read pairs per sample) or 12 Gb/sample (40 million read pairs per sample) *1 |
| Services | Sample quality check, library preparation, sequencing, and data analysis |
- *1 Data amount can be customized in 1 Gb increments. Please contact us for specific requirements.
Data Analysis
- Data Pre-Processing
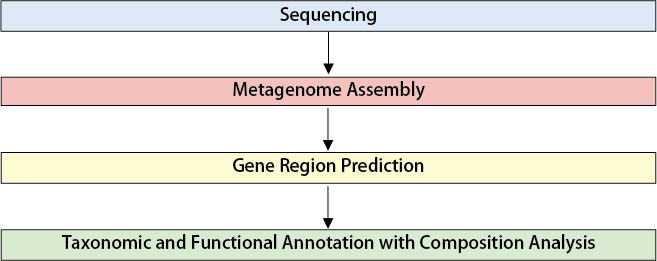
Raw data is subjected to quality control and host data removal to obtain usable data. - Metagenome Assembly
Reads are assembled using programs optimized for metagenomic analysis, reconstructing genomic fragments of organisms in the sample. - Gene Prediction and Abundance Analysis
Using MetaGeneMark, genes and protein-coding regions are detected. Redundant genes are removed to build a gene catalog. - Taxonomy Annotation (Micro NR)
Genes in the metagenome are classified to their respective species using the Micro NR database, enabling visualization of species composition and distribution in the sample. - Function Annotation (KEGG / eggNOG / CAZy)
Functions of genes are predicted by evaluating similarities with existing databases, including KEGG, eggNOG, and CAZy.
Sample Requirements
| Library Preparation | Sample Type | Total Amount | Concentration | Volume |
| PCR-Plus | DNA | ≥ 500 ng | ≥ 10 ng/µL | ≥ 30 µL |
| PCR-Free | ≥ 4 µg | ≥ 50 ng/µL | ≥ 30 µL |
