- HOME
- Products & Services
- Application Selection Guide
- Library Prep for Challenging DNA Samples
Library Prep for Challenging DNA Samples
Overview
Conventional DNA-Seq analysis relies on ligation-based adapters using DNA ligase, which is optimized for double-stranded DNA. This makes library preparation for single-stranded DNA (e.g., ssDNA viruses) or heavily degraded DNA (e.g., ancient DNA with multiple nicks) highly challenging.
Our service employs a specialized library preparation kit tailored for single-stranded DNA, enabling efficient library preparation for previously difficult-to-analyze samples.
This includes low-yield samples in the picogram range, making it ideal for samples that could not pass quality control due to factors such as DNA yield and degradation.
Applications
- Genome analysis of ssDNA viruses
- Sequencing damaged DNA from FFPE samples
- Sequencing ancient DNA
- Analysis of low-yield DNA such as cfDNA
- Sequencing heavily degraded or damaged DNA
Sample Requirements
| Total Amount | Concentration | Volume | Notes |
| ≥ 1 ng | ≥ 50 pg/µL*1 | ≥ 20 µL | DNA fragments ≥ 40 bp*2 |
The above concentration guideline is based on internal testing.
Important Notes
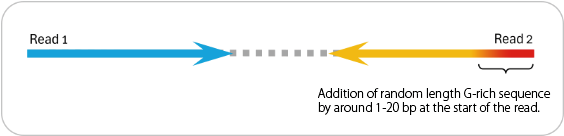
Due to the library preparation kit used in this service, Read 2 will include a random G-rich sequence of variable length at the start of the read.
This sequence must be trimmed during data analysis, which may reduce the effective read length.
Related Services
This service is available as an option for Illumina DNA-Seq.
Please request this service in combination with various applications compatible with Illumia sequencers (NovaSeq / NextSeq / MiSeq).
Back to "Products & Services"