- HOME
- Products & Services
- Order Procedure
Order Procedure
How to Order
-
STEP 1Fill in the Order Form
Download the order form for the desired service from the order form download page and fill in the required details.

If difficult sequences such as GC rich regions or complex secondary structures are anticipated, please indicate this in the remarks column of the order form.
-
STEP 2Send the Order Form (via E-mail)Attach the completed order form and send it via E-mail to seq@hssnet.co.jp .
Please refrain from ordering via FAX as the characters may be unclear. -
STEP 3Send SamplesInclude the printed order form with your samples and send them.
For details on sample packaging and shipping, please refer to the next section.
Note that the shipping address varies depending on the order content.For Sequencing Only or
Including PCR Product Purification
/ Primer WalkingAddress:Tokyo Haneda
1-6-2 Haneda Airport, Ota-ku, Tokyo
Japan 144-0041
Daiyon Sogo Building
Hokkaido System Science Co., Ltd.
Haneda Oligonucleotide Manufacturing Center, Sequencing Team
TEL:03-6840-1005For Orders Including DNA Extraction
or PCR Amplification
/ Microbial IdentificationAddress:Hokkaido Sapporo
1-2-1 Shinkawa Nishi 2-jo, Kita-ku, Sapporo
Japan 001-0932Hokkaido System Science Co., Ltd.
Analysis Team, Sequencing Division
TEL:011-768-5901 -
STEP 4Order Confirmation (Receipt Confirmation and Turnaround Time Notice)
* Same-day sample receipt is until 3:00 PM.After receiving the samples, we will confirm receipt and inform you of the estimated delivery time via E-mail.
To prevent delivery issues, please contact us if you do not receive a receipt confirmation from us.
Shipping Method
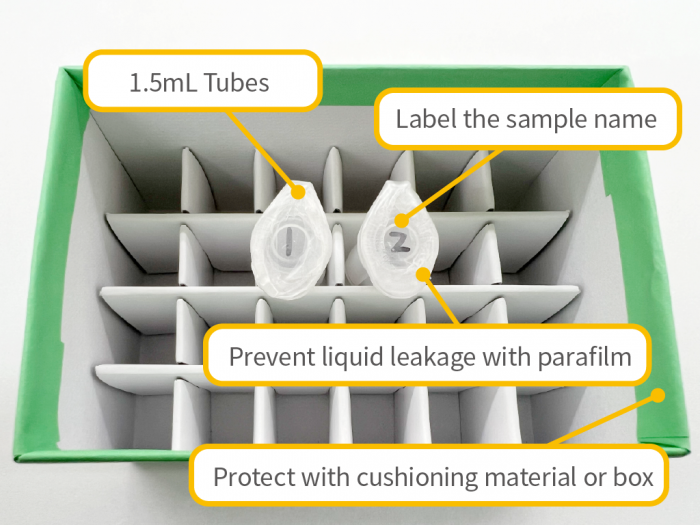
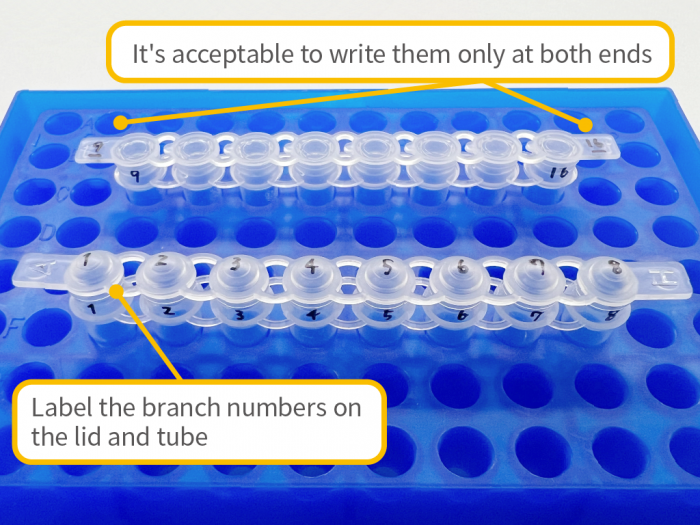
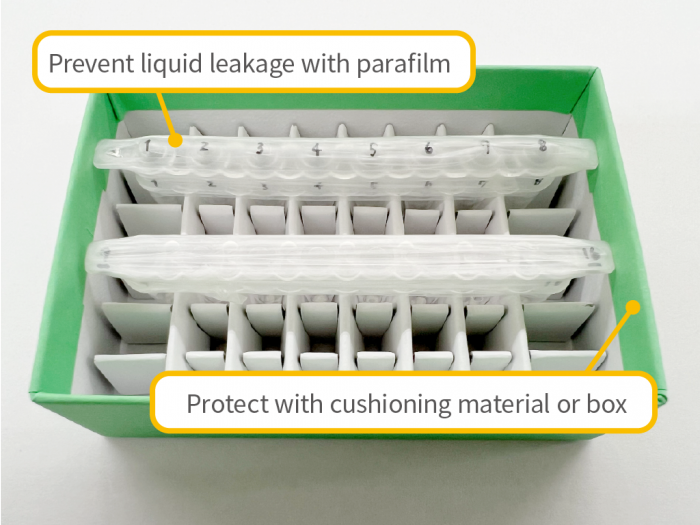
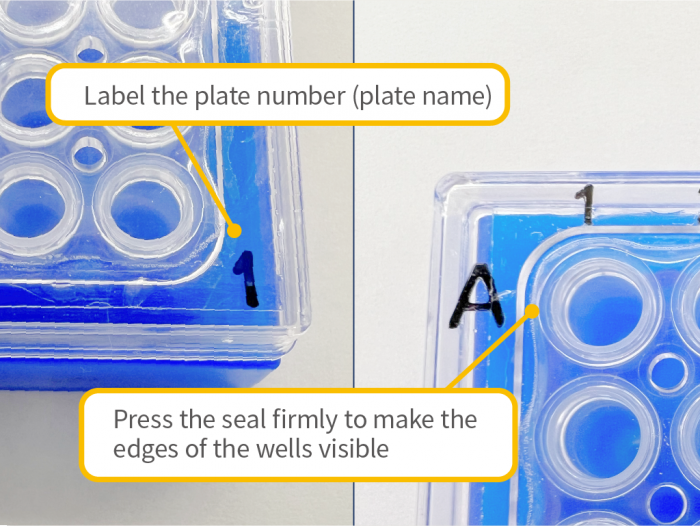
Sample Shipping Precautions
- – To prevent accidental damage or contamination (e.g., cracked or broken tubes, loose caps), please ensure samples are properly protected during shipping. We are not responsible for losses or damages that occur during transport.
- – Please ensure that the samples arrive on weekdays between 9:00 AM and 3:00 PM. We do not accept deliveries on weekends or public holidays.
- – Samples and primers used in sequencing analysis will be discarded after being stored for 3 business days following the completion of the analysis.
- – The shipping cost is the customer’s responsibility.
However, if you order eight or more analyses at once, you can use either [1] any delivery service with freight collect or [2] our designated Nippon Express shipping label (shipping cost covered by us). Please contact us if you need the Nippon Express shipping label.
Notes on Using Sequencing Analysis
Please understand the following points regarding the characteristics of sequencing analysis.
- Reading Range
- Due to the nature of capillary electrophoresis, the beginning of the read (approximately the first few tens of bases from the 3′ end of the primer) has poor resolution and is difficult to analyze. As a result, the sequence of the primer used in sequencing cannot be obtained as data.
- Factors Affecting Data Quality
- The success rate of sequencing analysis depends on the DNA sample’s concentration, purity, and base composition. Please note that special sequences (e.g., homopolymers, polyA, polyT, GC-rich regions, repeats) may result in poor or no data. It is challenging to obtain good data following homopolymers like polyT.
- Annealing Temperature
- Cycle sequencing reactions are typically annealed at 50°C. When providing custom primers, please be mindful of the primer’s Tm value.
- Using Mixed Primers
- Using mixed primers can lower the success rate of sequencing analysis. Variability in the molecular weight of cycle sequencing reaction products can reduce the accuracy of base calling.
Cautions
- – Product specifications and service details are subject to change without notice.
- – This service is intended for research purposes only and should not be used for purposes such as manufacturing or quality control of pharmaceuticals or food products, or for medical diagnostics.
- – We are not responsible for any losses or damages resulting from the use of this service.
- – Samples provided for analysis should comply with the P1 or P2 level regulations of the Cartagena Law on the use of genetically modified organisms by the Ministry of Education, Culture, Sports, Science, and Technology.
- – We do not accept pathogenic bacteria, infectious virus particles, etc. Samples should be prepared to the DNA state.
- – Please ensure informed consent has been obtained for human clinical samples.