- HOME
- Products & Services
- Evaluation of Off-target Effects
Evaluation of Off-target Effects

Service Description
Oligonucleotide therapeutics that target RNA, such as ASOs, siRNAs, and miRNAs, may produce unintended effects (off-target effects) by cross-hybridizing to sequences similar to their intended targets.
This service supports off-target evaluation in two phases:
(1) searching for candidate off-target genes for the specified sequence
(2) gene expression analysis using Microarrays or RNA-Seq
This service can help improve the safety and efficacy of oligonucleotide therapeutics.
Service Details
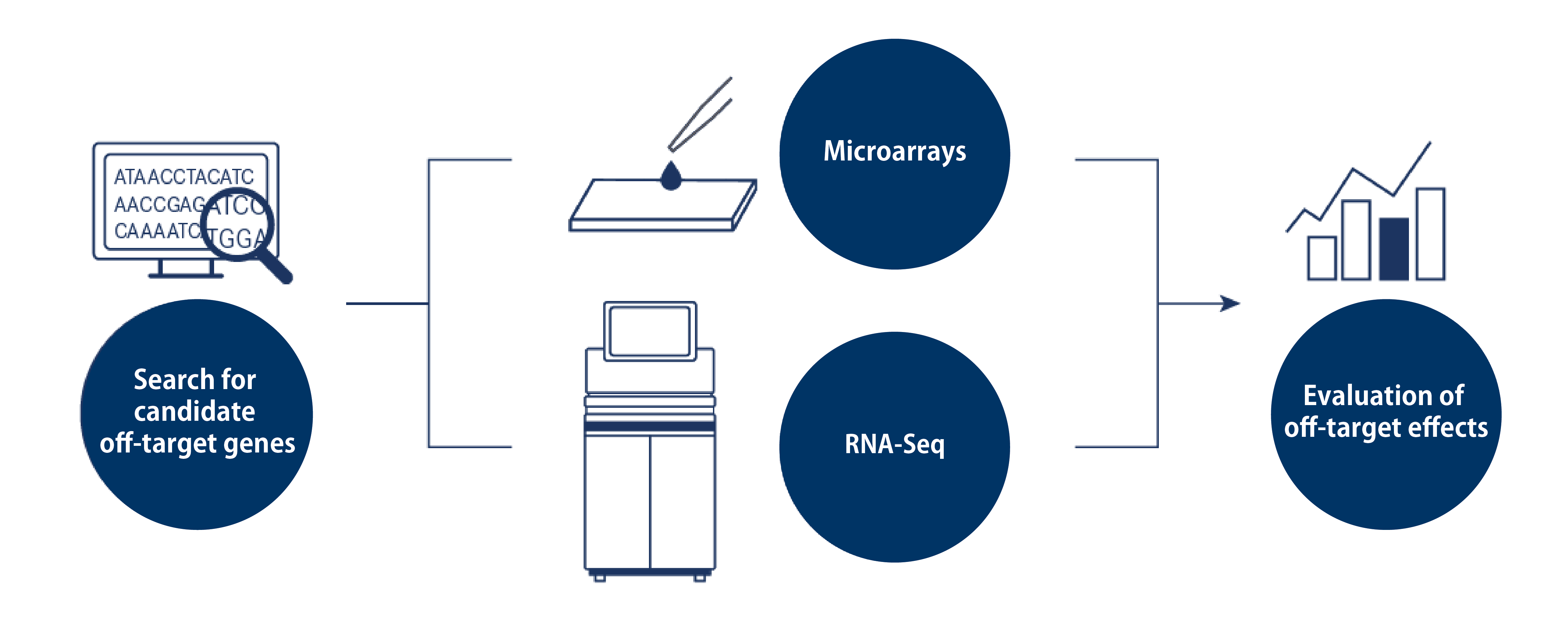
Using a dedicated sequence search tool*, we identify candidate genes that are likely to be affected as off-targets based on the sequence information you provide.
- *A high-speed nucleotide sequence search tool, “GGGenome,” is used.
Comprehensive gene profiling is performed using Agilent microarrays or RNA-Seq on an Illumina sequencer.
The resulting gene expression data are compared with the list of off-target candidates, and the suppression levels for each gene are plotted in a graph.
Delivery Data
The following data will be generated in addition to the standard delivery data from Microarrays / RNA-Seq.
| Data | format | Description |
|
・List of off-target candidate genes
|
txt / xlsx | List of candidate off-target genes from oligonucleotide therapeutic sequences identified using GGGenome |
|
・Visual summary of off-target evaluation
|
html | Graphs showing the expression behavior of candidate off-target genes (MA plot and cumulative frequency distribution) See figures below |
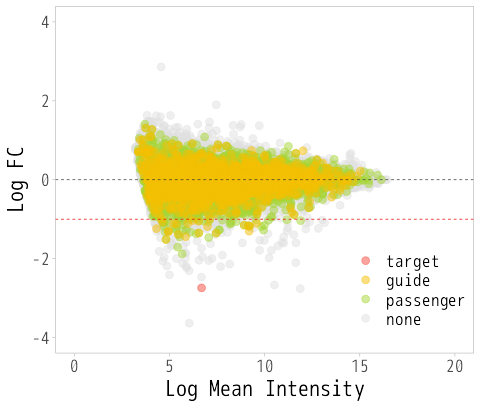
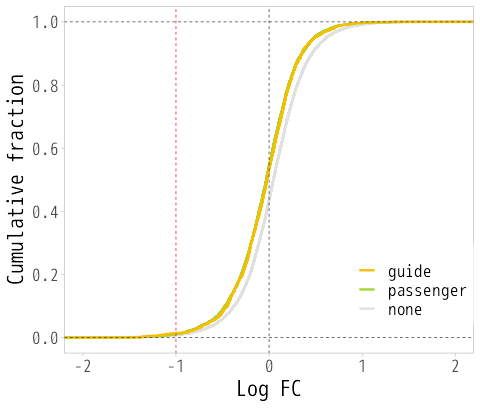
Figure: MA plot (left) and cumulative frequency distribution (right)
Required Sample
| Microarrays | RNA-Seq | |
| Total RNA | Total amount: 500 ng or more Concentration: 50 ng/μL or more Liquid amount: 10 μL or more * Dissolve in Nuclease-free water. * Spin-column type purification kit is recommended.(e.g. QIAGEN RNeasy series) |
Total amount: 600 ng or more Concentration: 20 ng/μL or more Volume: 30 μL or more * Dissolve in nuclease-free water. |
MA plots and cumulative frequency distributions are based on comparisons between samples, so data from a minimum of two samples are required.
It is common to use a mock-transfected sample as a control, but comparisons can be made not only with controls, but also between test samples themselves.
How to order
Order Flow
-
- 1Inquiry
- Please inquire from an Inquiry Form.
We will start to discuss with you about experimental design and analysis method.
-
- 2Quotation
- After confirming your order details, we will provide you a quotation and an order form to fill out.
-
- 3Fill and send the order form
- Please fill the order form and send it to us by e-mail.
-
- 4Sample submit
- Please ship out your sample to be arrived at us during week days.
- For details about how to ship, please see the below section “How to ship samples”.
How to ship samples
Please send your RNA samples with enough dry ice to keep them frozen until they arrive.
Please include the printed order form and send the package to the address below.
Sample shipping to:
Attn. Analysis Team
Hokkaido System Science Co., Ltd.
2-1, Shinkawa Nishi 2-1, Kita-ku
Sapporo, Hokkaido, 001-0932
TEL: 011-768-5903
- * Please inform us of the below information by E-mail upon sample shipping:
- 1. Estimated arrival date
- 2. Carrier
- 3. Tracking No.
- * We are afraid that we do not receive shipments on Saturdays, Sundays and holidays. Please make sure to arrange the shipments to be delivered to us during our operating hours (from 9:00 to 18:00) on weekdays.
