- HOME
- Products & Services
- Application Selection Guide
- Metatranscriptome Sequencing
Metatranscriptome Sequencing
Overview
Metatranscriptome sequencing generates RNA-Seq libraries from total RNA extracted from environmental samples containing diverse microorganisms.
This method reveals the microbial species present in the sample and the expression profiles of functional genes. Efficient analysis is achieved by removing ribosomal RNA using Ribo-Zero reagents.
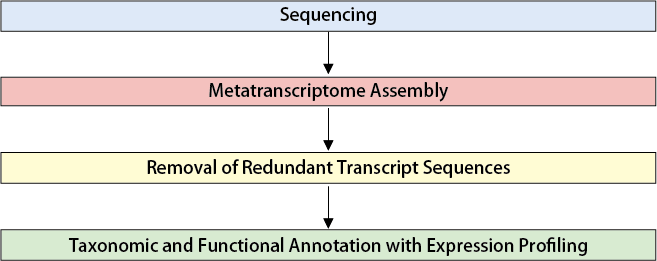
Workflow
Specifications
| Platform | Illumina NovaSeq 6000 |
| Read Method | Paired-End (150 bp x 2) |
| Data Amount | 6 Gb/sample (20 million read pairs per sample) or 12 Gb/sample (40 million read pairs per sample) *1 |
| Services | Sample quality check, library preparation, sequencing, and data analysis |
- *1 Data amount can be customized in 1 Gb increments. Please contact us for specific requirements.
Data Analysis
- Data Quality Control
Raw data is processed to ensure quality and converted into usable data. - Transcriptome Reconstruction
Non-target RNAs (e.g., rRNA, tRNA) are removed using NCBI reference sequences, and remaining reads are assembled using Trinity software.Redundant sequences are further removed using CD-HIT-EST to generate a unique gene set (unigenes). - Species Annotation
Unigenes are compared with sequences from bacteria, fungi, archaea, and viruses to identify biologically meaningful alignments.Results are combined with gene abundance data for visualization across taxonomic levels. - Function Annotation (GO / KEGG / eggNOG / CAZy)
Functions of unigenes are predicted based on similarity to known sequences in databases such as GO, KEGG, eggNOG, and CAZy. - Gene Expression Analysis / Gene Expression Difference Analysis
Reads are mapped to the reference transcriptome constructed by Trinity, and gene expression levels are calculated. Differentially expressed genes are identified through normalization and comparisons between samples or groups.Differentially expressed genes are identified through normalization and comparisons between samples or groups. - Enrichment Analysis (GO / KEGG)
Associations between differentially expressed genes and known genes in databases like GO and KEGG are evaluated to identify functional enrichment.
Sample Requirements
| Sample Type | Total Amount | Concentration | Volume |
| Total RNA | ≥ 1.5 µg | ≥ 50 ng/µL | ≥ 30 µL |
