- HOME
- Products & Services
- Application Selection Guide
- Gene Expression Analysis (Reference-Based)
Gene Expression Analysis (Reference-Based)
Overview
In RNA-Seq, efficient mRNA analysis requires the removal of ribosomal RNA (rRNA), which constitutes the majority of total RNA.
We use Oligo-dT beads or the Illumina Ribo-Zero Plus kit to deplete rRNA from the total RNA samples you provide, enabling the comprehensive sequencing of mRNA.
Workflow
Recommended Data Amount
| Sequencing Specification | Illumina 150PE |
| Data Amount | 20 million read pairs per sample |
Data Analysis
Gene Expression Analysis (Standard Analysis)
We calculate gene expression levels (TPM values) from RNA-Seq data.
Standard analysis includes fold-change calculations between samples or groups.
Statistical significance testing is available as an optional service.
Features:
Example of Delivered Data:
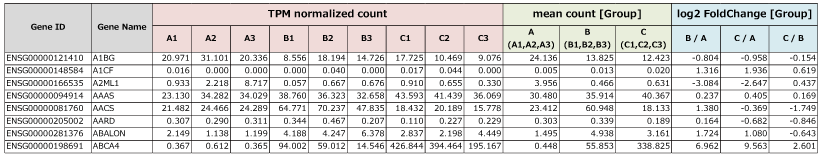
■ Gene Expression Table
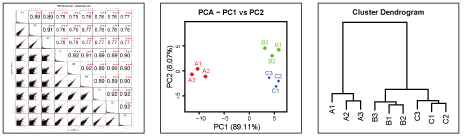
■ Various Figures
Visualization of expression profile comparisons between samples.
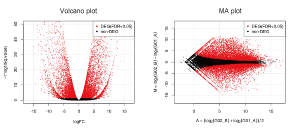
Statistical Significance Testing (Optional)
We perform statistical significance testing for gene expression changes between groups (recommended for n ≥ 3).
Example of Delivered Data:
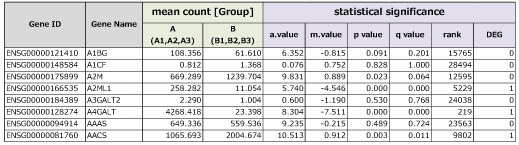
■ Statistical Test Results
■ Various Figures
GO Analysis (Optional)
We identify Gene Ontology (GO) terms highly associated with differentially expressed genes.
Example of Delivered Data:

Pathway Analysis (Optional)
We identify pathways closely associated with differentially expressed genes.
Delivery Data Example:
Sample Requirements
| Organism Type | Sample Type | Total Amount | Concentration | Volume |
| Eukaryotes | Total RNA | ≥ 600 ng | ≥ 20 ng/µL | ≥ 30 µL |
| mRNA | ≥ 100 ng | ≥ 3 ng/µL | ≥ 30 µL | |
| Prokaryotes | Total RNA | ≥ 1.5 µg | ≥ 50 ng/µL | ≥ 30 µL |
| mRNA | ≥ 100 ng | ≥ 3 ng/µL | ≥ 30 µL |
