- HOME
- Products & Services
- Antisense Oligonucleotide (ASO) Design & Synthesis
Antisense Oligonucleotide (ASO) Design & Synthesis
- Service Description
- Features
- Gapmer-type ASO
- Why Sequence Design Is Important
- Design Principle
- Order Flow
- Service Details
- Price and Domestic Lead-time
- Cautions
Veritas In Silico Inc. (VIS) and Hokkaido System Science Co., Ltd. (HSS) have a service agreement for antisense oligonucleotide (ASO) design and synthesis and jointly provide highly active gapmer-type ASO services using locked nucleic acids.
Service Description
A gapmer-type antisense sequence with locked nucleic acids introduced at both ends will be designed targeting your specified human gene.
VIS will perform the antisense sequence design, and HSS will synthesize the oligonucleotides.
- * The intellectual property right of the designed sequence will belong to the client and VIS.
Features
VIS has found that, in the translation knockdown process from mRNA to protein involving gapmer-type ASOs, the rate-limiting step is the binding of the antisense to the target mRNA. Furthermore, VIS has confirmed that targeting unstable regions in the mRNA is effective for accelerating this binding step by lowering the transition state energy.
Based on these findings, VIS designs ASOs by analyzing highly probable secondary structures within the target mRNA and selecting complementary regions, thereby providing ASOs with high efficacy and reduced off-target effects.
Gapmer-type ASO
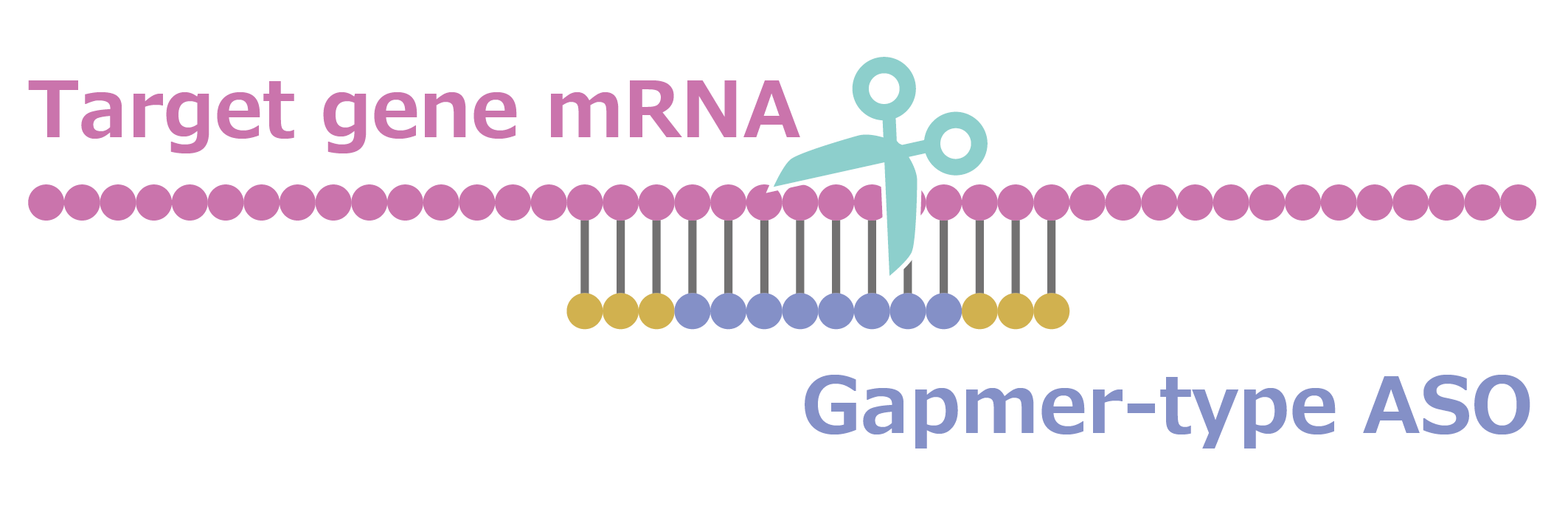
“Gapmer-type ASO” is a short DNA oligonucleotide that binds to mRNA of the target gene.
The mRNA/DNA duplex is cleaved by enzymes, thereby inhibiting the function of the target gene, which has led to the widespread use of these oligonucleotides as nucleic acid therapeutics.
Why Sequence Design Is Important
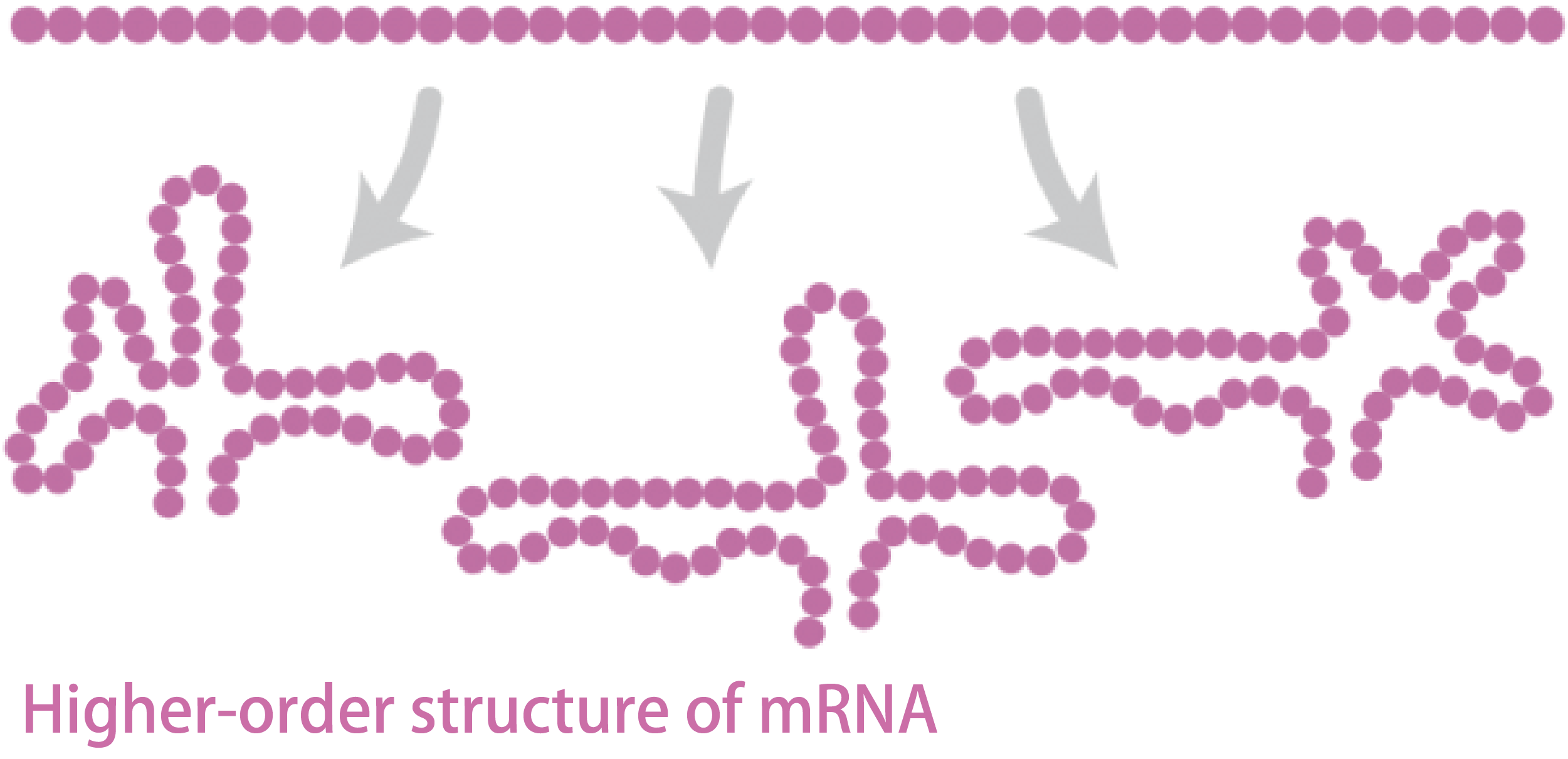
For ASOs to function efficiently, sequence design—deciding which region of the target mRNA to bind—is critical. This is because mRNA adopts higher-order structures in the cell, resulting in regions that are more or less accessible for binding.
Design Principle
Exploration of targetable regions and design of ASOs
| ● Step1 Prediction of mRNA structure The secondary structure of mRNA is predicted for frames of fixed base length using multiple software tools, accumulating all possible predicted structures (upper right figure). ● Step2 Structural analysis (MobyDick) The probability of existence is calculated for all predicted structures (large structures) in a frame. For each partial structure within a large structure, its probability is calculated as the sum of the probabilities of all large structures containing it (middle right figure). The frame is then shifted by three bases, and the prediction and analysis are repeated. ● Step3 Search for drug target sites (MobyDick) All predicted and analyzed partial structures are visualized in MobyDick, allowing their probabilities of existence to be readily assessed (bottom right figure). Potential druggable structures are explored based on their probabilities of existence, stability, structural characteristics, etc. ● Step4 Design of antisense sequences Antisense sequences are designed to target the explored druggable partial structures. |
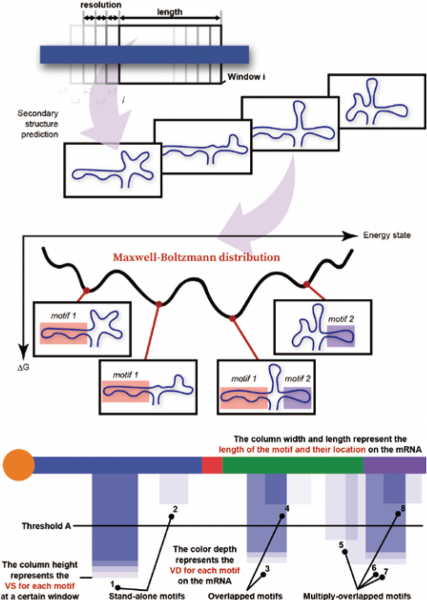 |
Order Flow
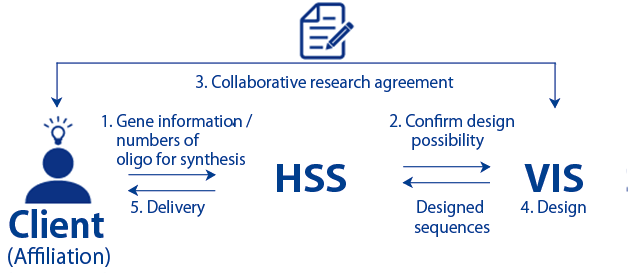
- 1. Please provide HSS with the information about the human genes for which you are requesting design, and the number of oligos required
- 2. HSS will confirm VIS about the design possibility for the requested gene.
- 3. If it can be designed, VIS will enter into an agreement with the client.
- 4. VIS will start to design.
- 5. HSS will synthesize the designed sequences and deliver the product to the client.
Service Details
- – Design service is available for human genes.
- – Based on the RefSeq ID or full-length mRNA sequence provided, we will use sequence design software built with a proprietary algorithm to design a gapmer-type antisense sequence with locked nucleic acids introduced at both ends.
- – The intellectual property rights for the designed sequences and oligonucleotides belong to the client and VIS.
- – Approximately 30 or 10 candidates of 14-base sequences will be provided. The sequences will be kept confidential when provided.
- – A sufficiently good sequence may be found from among the top 30 or more sequences; therefore, we recommend synthesizing all of the designed sequences.
- * If you should select less numbers of sequences due to budgetary constraints etc., please let us know how many from the top you would like to synthesize.
- – The synthesized oligonucleotides will be incorporated 3 bases of locked nucleic acid at both ends, and all bases will be phosphorothioate-modified.
- – The effectiveness of ASO designed using this service can not be guaranteed.
[Locked nucleic acid-containing Gapmer-type ASO]
| Synthesis Scale | 0.2 μmol scale | 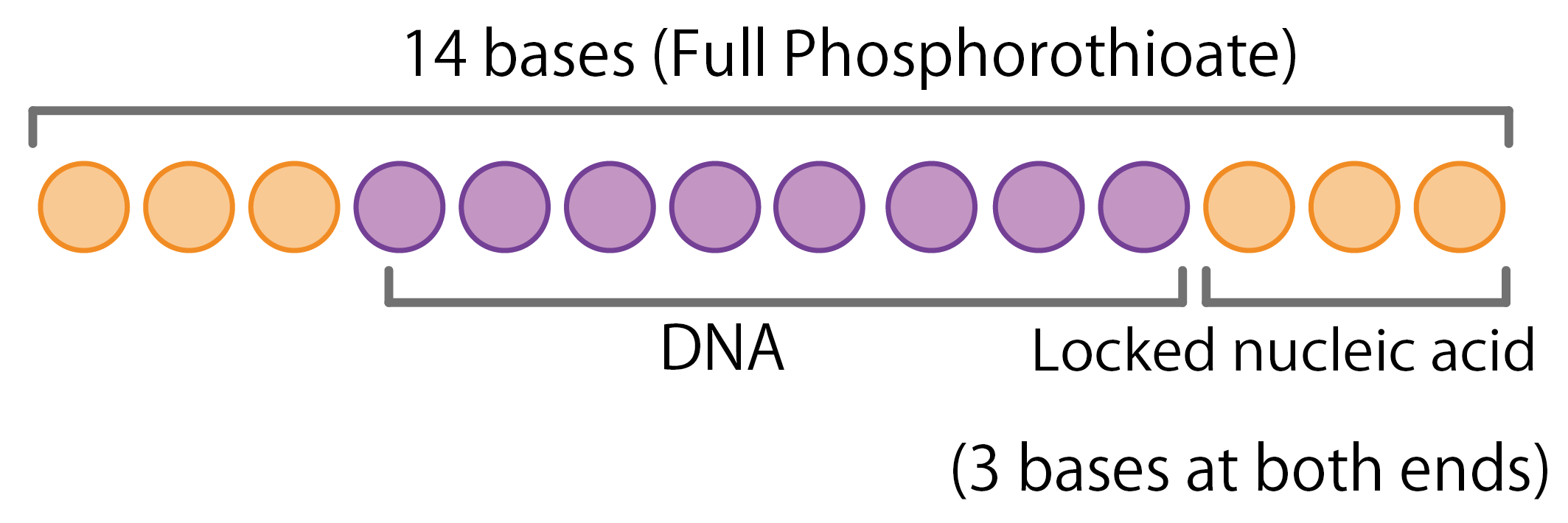 |
| Yield guaranteed | 1 OD | |
| Base length | 14 bases | |
| Modification | 3 bases at both ends: Locked nucleic acid All bases: Phosphorothioate (PS) |
|
| Purification | HPLC |
◆ Optional : Off-Target Search Results
We believe that the top priority in antisense design is to first identify an effective sequence. Once an effective sequence has been found, it is then possible to search for more preferable sequences that do not have off-target matches. (VIS’s design method inherently aims to avoid off-target effects when designing antisense sequences.) Therefore, during the initial design stage, we do not exclude any candidate sequences solely because they have potential off-target matches. However, as an option, we can additionally check the designed sequences for any perfect matches and provide the results as an attachment.
Price and Domestic Lead-time
- * Domestic lead-time varies depend on each order (design contents, numbers of synthesis etc.).
| Design Charge | Synthesis Charge | Option Off-Target Search Results |
Domestic lead-time | |
| Normal Plan | 100,000 / gene | 600,000 / 30 oligos (20,000 / oligo) |
375,000 | Design (VIS): approx. 2 weeks Synthesis (HSS): approx. 16 business days |
| Trial Plan only for academic clients |
50,000 / gene | 200,000 / 10 oligos (20,000 / oligo) |
Design (VIS): approx. 2 weeks Synthesis (HSS): approx. 12 business days |
Cautions
- – No orders can be cancelled due to the nature of the service.
For unavoidable cancellation, the fee for actual works will be charged. - – Product specifications and service contents may be changed without notice.
- – The base sequence information of designed sequences will not be made public.
- – The effectiveness of ASO designed using this service can not be guaranteed.
- – Due to the nature of this service, we are not responsible for any loss or damage that may arise from the results obtained through this service.
