- HOME
- Products & Services
- Application Selection Guide
- CRISPR Screening
CRISPR Screening
Overview
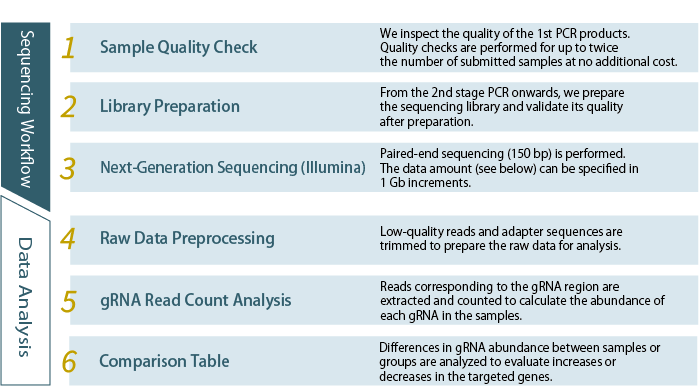
Our next-generation sequencing service supports genome-wide screening using CRISPR gRNA libraries. By providing PCR products of the gRNA region, we conduct deep sequencing and perform gRNA read count analysis.

Services
The data amount is calculated as follows:
Read Pair Count x Read Length x 2 (paired-end)
For example, sequencing 10 million read pairs at 150 bp:
10,000,000 x 150 bp x 2 = 3,000,000,000 b = 3 Gb
Coverage is determined by the following formula:
Read Pair Count / Number of gRNA Types = Coverage
For CRISPR screening analysis, coverage of approximately 100 – 500x is generally recommended.
For example, for a library with 120,000 types of gRNA at 100x coverage:
120,000 x 100 = 12 million read pairs, resulting in:
12,000,000 x 150 bp x 2 = 3.6 Gb
Please specify the desired data amount (read pair count) based on the type and desired coverage of your gRNA library.
Deliverables
| Item | Content | Format | Details |
| Reports |
・Work Report
|
Document | Overview of the workflow |
|
・Sequencing Report
|
Document | Details of the sequencing data | |
| Media – DVD-R – USB – HDD, etc. |
・Sequencing Data
|
fastq / txt / xlsx | Raw sequence data including corresponding quality scores (fastq) and a summary of sequencing results |
|
・Preprocessing Data
|
fastq / txt / xlsx | Data after quality control (QC) and trimming, along with a summary of the pre-processing steps | |
|
・gRNA Count Data
|
txt / xlsx | gRNA count data for each sample, calculated using the MAGeCK program (Example Data 1) | |
|
・Comparison Table
|
txt / xlsx | Comparison results of gRNA abundance between samples or groups, analyzed using the MAGeCK program (Example Data 2 / Example Data 3) | |
|
・Reference Data
|
txt | gRNA library sequence data used in the analysis | |
|
・Parameters
|
txt | List of programs and parameters used in the analysis | |
|
・Analysis Report
|
txt | Comprehensive documentation on data analysis | |
|
・Supplementary
|
txt | Guidance on free software for viewing fastq files and an overview of the library structure |
Example of Delivered Data: gRNA Read Count Table
| gRNA ID | gRNA Sequence | SampleA Count |
| ID_0001 | AAATACAAACAACTCTGAG | 213 |
| ID_0002 | GAAGTGTCCCAGGGCGAGG | 294 |
| ID_0003 | ACTCTATCACATCACACTG | 35 |
Example of Delivered Data: gRNA Count Comparison Table
| gRNA ID | Control | Test | LOG2FC | p_value | FDR | High in Treat | ||
|---|---|---|---|---|---|---|---|---|
| SampleA | SampleB | SampleC | SampleD | |||||
| ID_0001 | 213 | 274 | 883 | 175 | 1.12 | 0.9996 | 0.00699 | FALSE |
| ID_0002 | 294 | 412 | 1554 | 1891 | 2.29 | 0.0000 | 0.00000 | TRUE |
| ID_0003 | 421 | 368 | 566 | 759 | 0.75 | 0.9995 | 0.00726 | FALSE |
Example of Delivered Data: Positive/Negative Selection of Target Genes
| Gene | negative selection | positive selection | ||||
|---|---|---|---|---|---|---|
| p_value | FDR | Rank | p_value | FDR | Rank | |
| RPS5 | 0.000 | 0.000236 | 1 | 1.000 | 1.000 | 19147 |
| YARS | 0.000 | 0.000236 | 2 | 1.000 | 1.000 | 19146 |
| GTF2B | 0.000 | 0.000236 | 3 | 1.000 | 1.000 | 19145 |
| NRF1 | 0.000 | 0.000236 | 4 | 1.000 | 1.000 | 19144 |
| NLE1 | 0.000 | 0.000236 | 5 | 1.000 | 1.000 | 19143 |
Items to prepare

- 1. Prepare PCR products with linkers added using primers with our specified linker sequences (contact us for details).
- 2. The insert size (excluding linkers) should range between 200-400 bp, with the gRNA region located within the first 100 bp from the 5′ end. Design primers accordingly.
- 3. Submit PCR products with at least 5 ng (0.2 ng/µL or higher).
- 4. Purify PCR products to remove primers and nonspecific byproducts.
Day 0 (untreated) cells post-transfection
The gRNA library used before transfection
Provide the sequence information of the gRNA library. This can include:
Purchase details (URL or catalog number)
A list of IDs, sequences, and target genes
| gRNA ID | gRNA sequence | Gene |
|---|---|---|
| ID_0001 | AAATACAAACAACTCTGAG | GeneA |
| ID_0002 | GAAGTGTCCCAGGGCGAGG | GeneB |
| ID_0003 | ACTCTATCACATCACACTG | GeneC |
Download the appropriate form, complete the required details, and submit it.
Illumina Order Form (For CRISPR Screening, JPN)
